Advances in Medical Image Segmentation
This paper is a review on the advances in medical image segmentation technology over the past few years. With the increasing popularity of deep learning, there has been more innovation and application of these techniques in the medical space. Through an analysis of these approaches we can see the clear progression in innovation and the extensive applications.
- What is Medical Image Segmentation?
- V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
- TransUNet: Transformers make Strong Encoders for Medical Image Segmentation
- Segment Anything in Medical Images
- References
What is Medical Image Segmentation?
Medical image segmentation is a part of the medical image analysis process for images like CT scans and MRIs. It refers to the process of identifying pixels in medical images that correspond to organs, lesions, or other medical regions. The goal is to extract information about the shape and volume of organs, tissues, and other identified regions. Tackling this task with deep learning is a challenge as there is a high level of complexity and diversity with medical images [4]. This report will serve as a literature review of the advances in medical image segmentation over the past few years.
V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
At the time that this model was developed, deep learning was growing rapidly in popularity and CNNs were being applied to solve a variety of problems. However, a lot of the models were designed to work with 2D image slices, which results in a significant data loss for medical image segmentation tasks. The goal of V-Net was to be a CNN that was trained and developed to handle 3D image segmentation [3].
Implementation
V-Net is a fully convolutional neural network. The network is divided into stages that each have one to three convolutional layers. Convolutions are performed using kernels of the size 5x5x5. As stages progress, the path is compressed with 2x2x2 kernels being used (this decreases the feature map size). Downsampling is also used to reduce the size of the signals and increase the receptive field of the features. Instead of max pooling, V-Net uses strided convolutions [3]. This decreases memory usage and improves model interpretability. The model introduces a novel function, optimized using the Dice coefficient (measures similarity between datasets). This loss function was added to address the imbalance in foreground and background pixels that is commonly found in medical image segmentation tasks. The Dice coefficient measures the overlap between predicted and ground truth segmentations. This allows for the foreground and background to then be balanced.
\[D = \frac{2 \sum (p_i g_i)}{\sum (p_i^2) + \sum (g_i^2)}\]Training and Results
V-Net was trained on a dataset of prostate scans in MRI. Since there aren’t many annotated medical images available for training, the model was also trained on deformed versions of the training dataset.
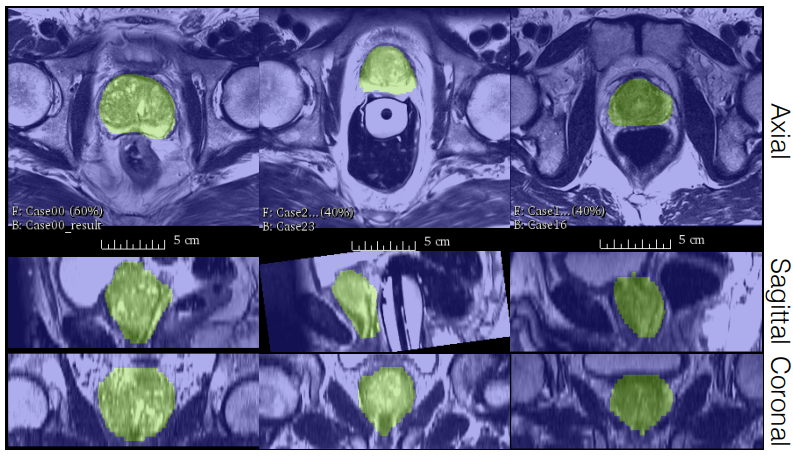
Fig 1. Image segmentation results on prostate MRI scans from the PROMISE 2012 dataset [3].
The trained model, V-Net, performs the image segmentation tasks on the dataset with a higher accuracy and less processing time than previous methods. The results can be seen in the table below where V-Net with Dice loss has the highest score.
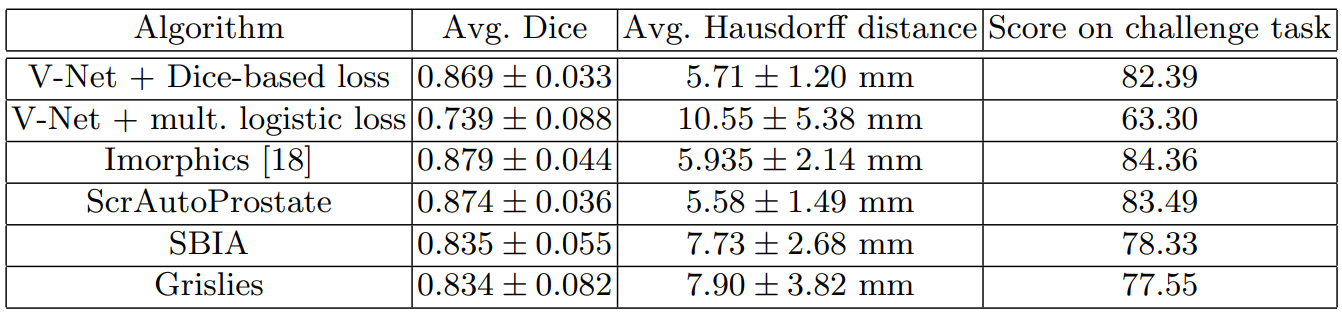
Table 1. V-Net’s performance on the PROMISE 2012 dataset compared to other methods [3].
TransUNet: Transformers make Strong Encoders for Medical Image Segmentation
A few years after the discussed work with CNNs in medical imaging, U-Net become a popular model for medical image segmentation. TransUNet takes a hybrid approach, taking parts of the U-Net and Transformer model architectures [1].
Implementation
U-Nets and CNNs have proven to be extremely useful for detail retention. A core limitation, however, is the inability to handle long-range relations. The transformer architecture addresses this with attention mechanisms that are designed to store long-range relations for sequence-to-sequence prediction. ResNet-50 is used first as a CNN encoder for initial feature extraction. Then a Vision Transformer (ViT) layer is used. This order allows high-resolution CNN feature maps to be leveraged and performs better than just using a pure Transformer for the decoder. After this, a cascade upsampler is used to output the final segmentation mask. Multiple upsampling blocks are cascaded until the full correct resolution is reached [1].
Training and Results
The model was trained on a multi-organ segmentation dataset called the MICCAI 2025 Multi-Altas Abdomen Labeling Challenge.
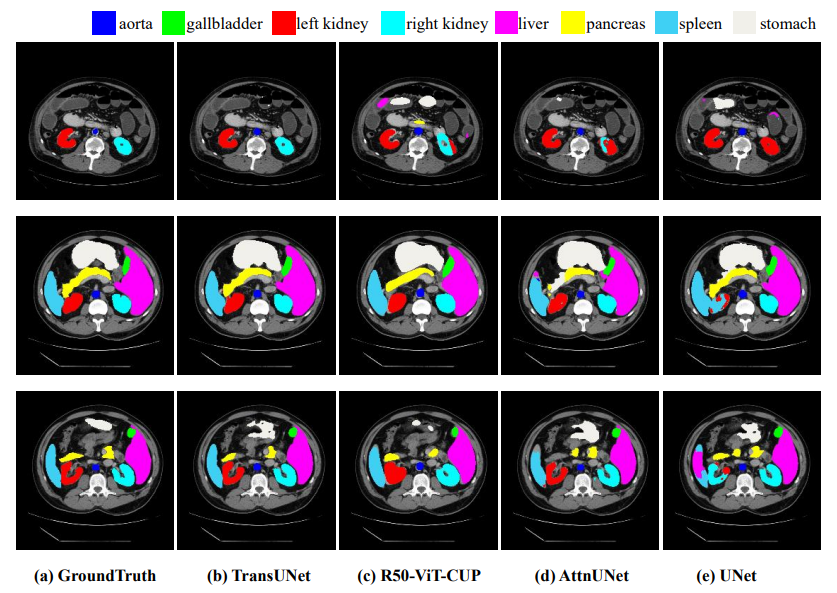
Fig 2. Comparison of results from different architecture approaches [1].
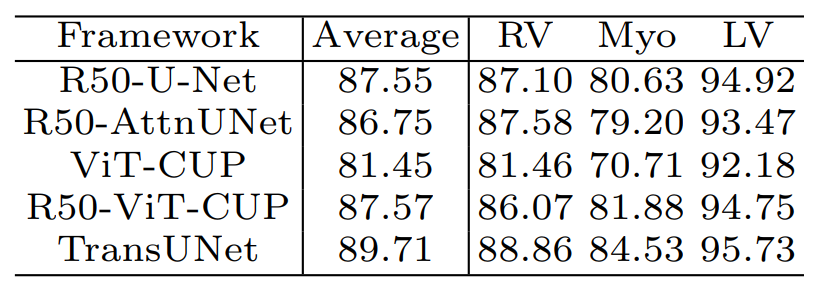
Table 2. TransUNet’s performance on the dataset compared to other methods [1].
Segment Anything in Medical Images
Up until this point, many of the models and advances in medical image segmentation were focused on developing models that needed to be tailored towards specific modalities or conditions. However, in practice, it is valuable to have generalizability across different types of medical image segmentation tasks. MedSAM addresses this as a model for universal medical image segmentation [2].
Implementation
The model is based on the vision transformer architecture. The model was initialized with a pre-trained SAM model and masked autoencoder modeling. Then it was trained with fully supervised training. The loss function is a combination of the Dice loss and cross-entropy loss. The AdamW optimizer was used for parameter optimization. MedSAM is also unique with a promptable interface. Compared to the previous automatic segmentation processes, MedSAM allows users to identify targets with bounding boxes [2]. This is an important feature for handling multiple types of medical images.
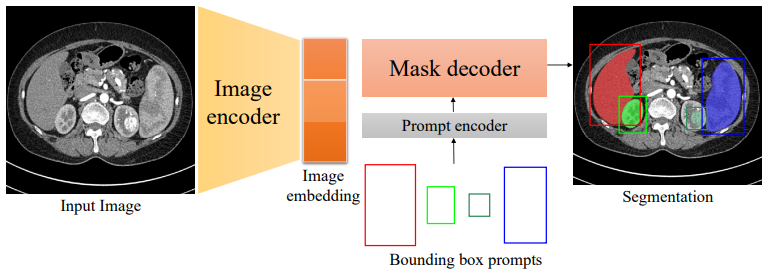
Fig 3. A diagram of the MedSAM architecture, including the prompt encoder [2].
Training and Results
One of the major issues that the other specialist models had was the lack of data availability for training. The researchers working on MedSAM curated their own set of images and 3d datasets from public sources. Images were converted and normalized to ensure uniformity and compatibility. MedSAM performance is on par with specialist models, even though it works with a much larger range of medical images.
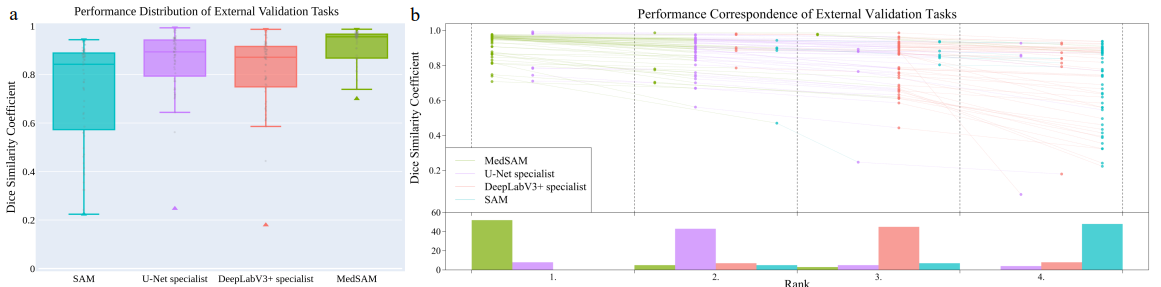
Fig 4. MedSAM performance compared against SAM, U-Net, and DeepLabV3 [2].
References
[1] Chen, Jieneng, et al. “TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation”. arXiv preprint arXiv:2102.04306 (2021)
[2] Ma, Jun, et al. “Segment anything”. arXiv preprint arXiv:2304.02643 (2023)
[3] Milletari, Fausto, et al. “V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation”. arXiv preprint arXiv:1606.04797 (2016)
[4] Yao, Wenjian “From CNN to Transformer: A Review of Medical Image Segmentation Models”. arXiv preprint arXiv:2308.05305 (2023)