Toward Unified Diffusion–MLLM Systems in Biomedical AI - A Survey of Integration Strategies and Research Directions
Diffusion models and multimodal large language models (MLLMs) have become pivotal to biomedical AI, excelling at generating high-quality medical images and interpreting clinical texts, respectively. Although these approaches have complementary strengths, their integration in biomedical applications remains limited. This review systematically analyzes over 20 studies that employ diffusion and MLLM techniques for tasks such as medical image synthesis, report creation, visual question answering, and cross-modal retrieval. We highlight emerging general-domain integration frameworks that offer promising approaches toward closer integration. Based on this analysis, we propose a taxonomy of four integration approaches and evaluate existing biomedical systems across multiple key dimensions. Our findings reveal a persistent gap between current modular implementations and the unified architectures needed for seamless clinical reasoning. Finally, we outline critical obstacles related to data fragmentation, architectural design, clinical validation, and evaluation protocols; we suggest research avenues to advance integrated foundation models for end-to-end multimodal reasoning within clinical workflows.
- Introduction
- Background
- Current Biomedical Approaches
- Toward Integration: A Proposed Taxonomy
- Technical Challenges and Research Directions
- Future Directions for Integrated Diffusion–MLLM Systems
- Conclusion
- References
Introduction
Diffusion models and large language models (LLMs) each substantially address distinct aspects of clinical reasoning and representation in biomedical AI. Diffusion models such as denoising diffusion probabilistic models (DDPMs) [8] and latent diffusion models (LDMs) [18] have demonstrated strong performance in generating high-resolution medical images and segmenting anatomical structures [9, 27, 28]. Meanwhile, LLMs such as GPT-4 [17] and BioGPT [14] have revealed their effectiveness in clinical text generation, report summarization, and medical question answering [11, 19].
Despite their complementary capabilities, these models are typically deployed in isolation. Diffusion models often lack task-specific semantic grounding, while LLMs are not inherently designed to process visual inputs. This separation limits their utility in clinical workflows that require joint reasoning over medical images and text [21, 23].
This paper surveys the current state of diffusion and multimodal LLM (MLLM) approaches in biomedical applications, focusing on over 20 recent studies across core tasks such as image synthesis, report generation, question answering, and retrieval(Figure 1). While truly integrated systems remain rare in this domain, emerging general-purpose architectures (e.g., LLaDA, DiffusionGPT, and NExT-GPT) demonstrate viable integration paradigms. Building on these examples, we propose a taxonomy of integration strategies suited to clinical settings and outline key challenges in data alignment, model design, and clinical validation. Our goal is to provide a foundation for advancing unified diffusion–MLLM systems in biomedical AI.
Background
Diffusion Models for Medical Imaging
Diffusion models generate data by means of a progressive denoising process, starting from pure noise. The DDPM framework [8] learns to reverse a Markov chain of noise additions to produce samples from a learned data distribution. Latent diffusion models (LDMs) [18] extend this by operating in a compressed latent space using a pre-trained autoencoder. A U-Net performs denoising within the latent space, and a decoder reconstructs high-resolution outputs. Stable Diffusion, a popular LDM, incorporates cross-attention with text embeddings to guide generation [5, 18].
In biomedicine, diffusion models have achieved success in generating synthetic MRIs, chest X-rays, and histopathology slides [9, 27, 28]. They are especially useful for data augmentation and modeling rare pathologies, in situations where real-world data is limited. Despite their versatility, a key limitation remains computational efficiency, particularly for 3D imaging [4, 21]. Ongoing work focuses on faster sampling and lightweight model architectures.
Multimodal Language Models in Biomedicine
Multimodal large language models (MLLMs) extend traditional LLMs to jointly process language and visual inputs. Early systems relied on late fusion, where visual features extracted from CNNs were combined with text embeddings at the output stage [3]. This design facilitated multimodal classification but offered limited interaction between modalities. More recent approaches use mid-fusion strategies, integrating image tokens into the language model via cross-attention. Flamingo [1] and LLaVA [13] exemplify this design and have been adapted to biomedical tasks through variants such as Flamingo-CXR [21] and LLaVA-Med.
Generative MLLMs extend these capabilities further by supporting not only understanding but also synthesis. Models like MedM2G [4] exhibit joint generation across modalities using structured prompts or image tokens as input. These architectures support data generation, simulation of rare cases, and augmentation for low-resource clinical settings [2].
Integration Paradigms from General AI Research
While most biomedical applications deploy diffusion models and language models independently, recent work in general-domain AI has introduced architectures that integrate these components. These systems offer promising templates for unified generation and multimodal reasoning.
LLaDA [16] replaces the standard autoregressive decoder with a diffusion-based alternative for language modeling. By learning to denoise corrupted text representations, it reveals that diffusion models can be competitive in natural language generation, extending their use beyond vision.
DiffusionGPT [25] enables LLMs to control image generation via structured conditioning and semantic layout prompts, showing that text-guided diffusion can be modular and interpretable.
NExT-GPT [26] generalizes this further by unifying vision, text, audio, and video through shared latent embeddings and a multimodal diffusion decoder. It highlights that scalable, any-to-any multimodal generation is feasible within a unified architecture.
These approaches, while developed outside the biomedical domain, illustrate integration strategies that may be adapted for future clinical systems.
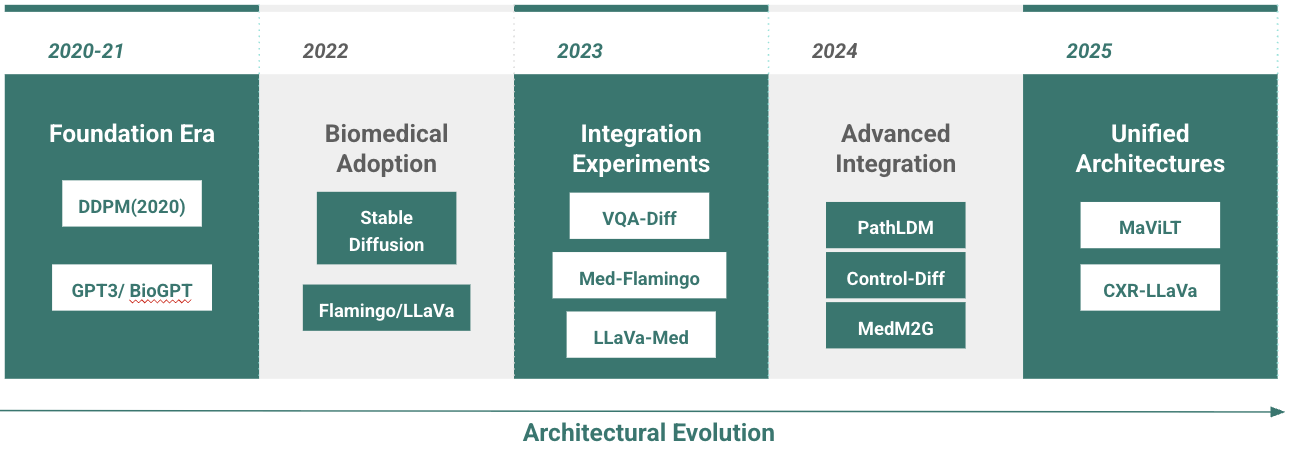
Current Biomedical Approaches
While diffusion models and MLLMs have independently advanced biomedical AI, integrated architectures remain rare. In particular, the combined potential of diffusion models’ precise generation and MLLMs’ advanced reasoning remains underutilized. In this section, we examine recent medical imaging and language processing systems through the lens of future integration, grouping them into three categories: diffusion-based pipelines, MLLM-centric approaches, and emerging efforts toward architectural fusion.
Diffusion-Based Medical Applications
Diffusion models have been increasingly applied to biomedical tasks due to their capacity for generating high-fidelity, diverse samples under limited supervision. Several recent systems demonstrate both the adaptability and clinical relevance of these models in image and text synthesis tasks.
ControlDiff [22] is a non-autoregressive radiology report generation framework that addresses the issue of semantic drift in autoregressive decoding. It introduces a task-specific noise generator (TNG), combining a global component based on common visual features and n-grams, with a local component derived from under-detected regions and rare tokens. The combined noise guides a diffusion decoder conditioned on visual inputs, improving clinical precision. On MIMIC-CXR, ControlDiff achieves a BLEU-4 of 0.132 ± 0.003, significantly outperforming both Transformer and vanilla diffusion baselines.
CoDiXR [15] adapts composable diffusion to multi-modal biomedical generation by training three modality-specific latent diffusion models for frontal view, lateral view, and report generation. These are aligned via a contrastive latent space using InfoNCE loss. On MIMIC-CXR, CoDiXR achieves a Fréchet Inception Distance (FID) of 0.86 and BLEU-4 of 0.22, with pathology AUROC ranging from 0.84 to 0.91, demonstrating strong cross-modal consistency.
PathLDM [27] presents the first text-conditioned latent diffusion model tailored to histopathology. Conditioned on distilled pathology reports using a GPT-based encoder, PathLDM outperforms prior models with a FID of 7.64 on the TCGA-BRCA dataset, compared to 30.1 for the closest text-conditioned baseline. Architectural modifications to the encoder, U-Net, and conditioning mechanisms contribute to this performance.
DiffBoost [28] proposes edge-guided, text-conditioned diffusion for medical image synthesis aimed at segmentation augmentation. It improves segmentation accuracy across multiple modalities by generating anatomically plausible images from sparse supervision. Examples include breast ultrasound (+13.87%), spleen CT (+0.38%), and prostate MRI (+7.78%). It is among the first frameworks to explore text-guided diffusion specifically for segmentation.
MedM2G [4] introduces a unified multimodal diffusion framework for medical generation across CT, MRI, and X-ray, supporting tasks including text-to-image, image-to-text, and cross-modality synthesis. Through cross-guided latent flows and modality-invariant alignment, MedM2G achieves state-of-the-art results across five generation tasks on ten datasets, highlighting its generalizability and clinical relevance.
Together, these models provide a foundation for integrating diffusion-based synthesis into multimodal biomedical systems, offering performance, controllability, and semantic grounding. However, they lack the sophisticated reasoning capabilities that MLLMs provide, creating clear opportunities for architectural integration.
MLLM-Based Medical Applications
Multimodal Large Language Models (MLLMs) are increasingly prominent in biomedical AI, offering strong performance across visual question answering, report generation, and retrieval-based inference. Recent work fine-tunes general-domain architectures for specialized medical domains, adapting them through multimodal supervision, domain-specific pretraining, and retrieval augmentation.
General Biomedical Vision–Language Models
LLaVA-Med [13] builds on the LLaVA framework by aligning biomedical figure–caption pairs extracted from PubMed Central and using GPT-4 to generate instruction-following data. A two-stage curriculum first tunes biomedical vocabulary, then adapts the model for open-ended conversations. Trained on 8 A100 GPUs in under 15 hours, LLaVA-Med achieves 50.2% of GPT-4 performance on multimodal VQA tasks and outperforms the base LLaVA model across diverse biomedical domains including CT, MRI, and histology.
Med-Flamingo [21] is a few-shot extension of the Flamingo model evaluated on VQA-RAD and PathVQA. It improves clinical evaluation scores by ~20% over baselines. On pathology datasets where models tend to underperform, Med-Flamingo still yields superior BERT-sim and exact match metrics, validating the few-shot adaptability of vision–language systems for medical QA.
BiomedCLIP [3] enhances CLIP by replacing GPT-2 with PubMedBERT for the text encoder and adapting Vision Transformers to better capture biomedical image structure. It introduces patch dropout and custom tokenization, yielding stronger image–text retrieval performance compared to PubMedCLIP and MedCLIP on radiology benchmarks.
Specialized Clinical Applications
CXR-LLaVA is a large-scale multimodal model for chest X-ray interpretation, combining a vision transformer trained on 374,000 labeled CXRs with a language decoder adapted from LLaVA. Fine-tuned on 217,000 report-paired CXRs, it achieves an F1 of 0.81 on internal and 0.62 on external pathology benchmarks, surpassing GPT-4-Vision and Gemini-Pro-Vision. In human evaluation, its autonomous reports were rated acceptable in 72.7% of cases, approaching the 84% threshold of gold-standard reports.
Foundation Model Approaches
Me-LLaMA is a family of open-source medical LLMs derived from LLaMA, trained on 129 billion tokens and 214,000 medical prompts. The 70-billion-token variant required over 100,000 A100 GPU hours and shows strong generalization on 12 benchmarks, surpassing ChatGPT and GPT-4 on 7 and 5 datasets, respectively. It supports complex diagnosis via instruction tuning and ranks comparably to GPT-4 on human-evaluated clinical case questions.
MAViLT [6] introduces a unified vision–language transformer using VQ-GAN tokenization and a clinical loss composed of reconstruction, gradient, and perceptual components. Pretrained with masked modeling and instruction tuning, it achieves a BLEU-4 of 0.486 and FID of 21.1 for CXR report generation and image synthesis, respectively. VQA accuracy on SLAKE reaches 68.5%, and radiologists score its reports a 4.5 out of 5 for completeness.
Collectively, these MLLM architectures push the frontier of clinical AI, demonstrating sophisticated multimodal reasoning capabilities. However, they operate independently from diffusion-based generators, lacking the controlled generation capabilities that diffusion models provide—a promising avenue for future integration.
The Integration Gap
Despite the parallel successes of diffusion models and MLLMs in biomedical AI, architectural integration between the two remains rare. A few early-stage approaches have explored pipeline-style fusion, but these fall short of deep, unified designs that tightly couple generative and reasoning capabilities.
Late Fusion via Diffusion-Based Visual Encoding. Bian et al. [2] propose a two-stage framework for medical VQA, using classifier-guided conditional diffusion to learn robust visual representations, followed by late-stage fusion with a GRU-based language model. The diffusion model is frozen after pretraining, and a semantic encoder guides sampling through gradient-based conditioning:
\[\hat{\epsilon}_{\theta,\phi}(x_t, t) = \epsilon_\theta(x_t, t) - \sqrt{1-\alpha_t} \nabla_{x_t} \log p_\phi(y|x_t)\]| where $\epsilon_\theta(x_t, t)$ is the noise prediction network, $p_\phi(y | x_t)$ is a classifier predicting labels from noised images, and $\alpha_t$ controls the noise schedule. While the model outperforms baselines on VQA-RAD and SLAKE, the integration remains shallow, with limited feedback between modalities. |
Retrieval-Augmented Pipelines. FactMM-RAG [20] exemplifies a retrieval-augmented multimodal generation pipeline. Instead of joint training, it conditions generation on reference reports selected by a multimodal retriever trained with RadGraph-derived factual pairs. Although this setup improves factual accuracy, the modular structure lacks architectural synergy between vision and language models. Similar retrieval-augmented methods like RAMM also treat components as loosely connected modules.
Diffusion–LLM Integration in General AI. Outside biomedicine, recent work indicates the feasibility of diffusion-based language modeling. For instance, LLaDA [16] trains a Transformer on masked token prediction via a forward masking and reverse denoising process, replacing the autoregressive paradigm. This suggests that tightly integrated diffusion–LLM architectures are possible, although biomedical applications have yet to adopt these innovations.
These examples illustrate a critical opportunity: existing approaches tend to use late or modular fusion with minimal architectural entanglement. Unified integration, where diffusion and multimodal reasoning components co-train or co-infer, remains largely unexplored in the biomedical domain. This gap motivates our taxonomy in Section 4, which systematizes current and emerging integration strategies.
Toward Integration: A Proposed Taxonomy
We organize upcoming efforts to combine diffusion models and multimodal language systems into four integration strategies,Figure 2. Each reflects a distinct architectural approach to aligning visual and textual reasoning in biomedical tasks. Our taxonomy highlights representative models, training methods, and technical gaps, aiming to guide future research toward more unified systems.
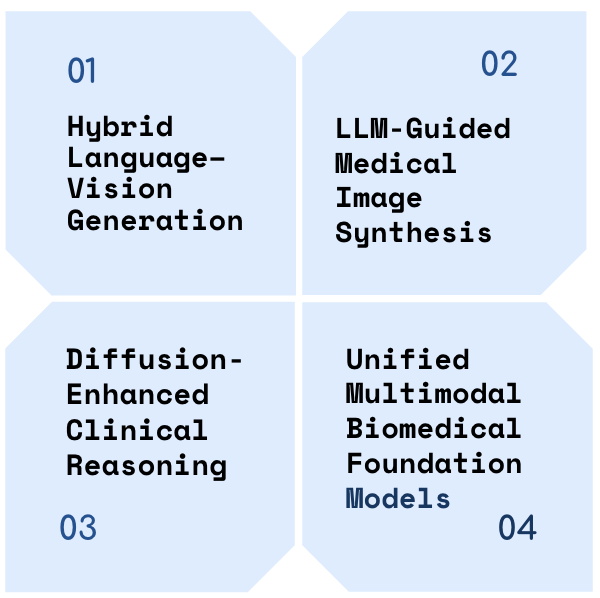
Hybrid Language–Vision Generation
Hybrid systems generate both medical images and text within a shared model. These architectures typically use token-based representations, enabling bidirectional synthesis between modalities.
MAViLT [6] exemplifies this strategy by combining VQ-GAN tokenization with clinical loss regularization to preserve diagnostic fidelity. The reconstruction objective integrates spatial and perceptual losses:
\[\mathcal{L}_{\text{recon}} = |x_v - \hat{x}_v|_1 + \lambda_{\text{grad}} \| \nabla x_v - \nabla \hat{x}_v \|_2^2 + \lambda_{\text{feat}} \| \phi(x_v) - \phi(\hat{x}_v) \|_2^2\]where $x_v$ is the original image, $\hat{x}_v$ is the reconstructed image, $\nabla$ denotes the gradient operator, and $\phi(\cdot)$ is a pretrained radiograph encoder. This enables both report-to-image and image-to-report generation. MAViLT achieves a BLEU of 0.486 for report synthesis and FID 21.1 for image generation on MIMIC-CXR.
In the general domain, LLaDA [16] replaces autoregressive decoding with a diffusion-based masking process. During training, a random portion of tokens is masked:
\[x_t \sim \text{Mask}(x_0, t), \quad t \sim U[0, 1]\]and the model learns to predict all masked tokens simultaneously in a single denoising step.
Despite their promise, current hybrid models are limited to single-domain datasets or unimodal training objectives. Full integration across diverse biomedical modalities remains an open challenge.
LLM-Guided Medical Image Synthesis
This category includes systems where language models serve as semantic controllers for medical image generation. These approaches enable diffusion models to generate domain-specific visuals guided by structured clinical prompts or embeddings.
PathLDM [27] uses a GPT-based encoder to process clinical descriptions, conditioning a latent diffusion model to synthesize histopathology images. It outperforms baseline models like DALL-E and LDM, achieving an FID of 7.64 compared to 30.1, while preserving text-image consistency under pathological constraints.
CoDiXR [15] trains three modality-specific latent diffusion models for frontal-view, lateral-view, and report generation, aligned through a contrastive latent space using InfoNCE loss. It supports compositional prompts and achieves an FID of 0.86 and BLEU-4 of 0.22 on MIMIC-CXR, enabling coherent generation across multiple modalities.
DiffBoost [28] enhances segmentation performance through text-guided image synthesis. A lightweight text encoder conditions diffusion to generate task-specific augmentations, leading to +13.87% IoU improvement on breast ultrasound and +7.78% on prostate MRI. This demonstrates the value of semantically aligned augmentation in low-data clinical tasks.
These models show the potential of coupling LLM-derived features with diffusion for guided generation. However, most systems adopt modular designs without unified embeddings or joint optimization across modalities.
Diffusion-Enhanced Clinical Reasoning
In this class of models, diffusion processes support or enhance the reasoning capabilities of language models, particularly for tasks like medical question answering and report generation. Unlike Section 4.2, where LLMs guide image synthesis, these systems use diffusion to refine or structure textual outputs.
VQA-Diff [2] learns medical visual representations via classifier-guided conditional diffusion, which are later fused with a GRU-based question encoder for visual question answering. The guidance signal modifies the denoising process as:
\[\hat{\epsilon}_{\theta,\phi}(x_t, t) = \epsilon_\theta(x_t, t) - \sqrt{1-\alpha_t} \nabla_{x_t} \log p_\phi(y|x_t)\]| where $\epsilon_\theta(x_t, t)$ is the noise prediction network, $p_\phi(y | x_t)$ predicts labels from noised images, and $\alpha_t$ controls the noise schedule. On VQA-RAD, the model achieves 74.1% accuracy, surpassing the CPRD baseline by 1.4%, with stronger performance on open-ended questions. |
ControlDiff [22] generates radiology reports by injecting clinical intent into the denoising process through a task-specific noise generator (TNG). The TNG combines global linguistic patterns and local medical cues:
\[\begin{align} \text{TNG} &= \text{Global}(\text{visual features, n-grams}) \\ &\quad + \text{Local}(\text{under-detected regions, rare tokens}) \end{align}\]This architecture improves report fidelity, achieving BLEU-4 of 0.132 (with a 0.003 error margin) on MIMIC-CXR, with measurable gains in n-gram diversity and entity accuracy.
FactMM-RAG [20] introduces a retrieval-augmented generation pipeline using RadGraph-based factual grounding. Retrieved reports are fused with image embeddings before generation, improving factual consistency without altering the diffusion architecture. It yields +6.5% F1CheXbert and +2% F1RadGraph over standard generation models.
These systems demonstrate early evidence that diffusion models can improve text-level clinical reasoning. However, architectural modularity and limited feedback loops still constrain deeper integration.
Unified Multimodal Biomedical Foundation Models
This category encompasses large-scale models designed to unify medical image and text modalities within a single architecture. These systems aim to support text-to-image generation, report synthesis, retrieval, and cross-modality understanding with shared embeddings and compositional interfaces.
MedM2G [4] introduces cross-guided latent diffusion flows to support text-to-image, image-to-text, and image-to-image generation across CT, MRI, and X-ray applications. By encoding modality-specific priors into a unified latent space and conditioning generation using cross-modal prompts, the model achieves state-of-the-art results across five generation tasks on ten public datasets.
Me-LLaMA [24] represents a language-first biomedical foundation model trained on 129 billion tokens with 214,000 medical prompts, requiring over 100,000 A100 GPU hours. Out of 8 biomedical NLP benchmarks, it outperforms ChatGPT on 7 and GPT-4 on 5, establishing it as a competitive language backbone for future multimodal fusion.
CXR-LLaVA [12] specializes in chest X-ray understanding, trained on 592,000 radiographs. It achieves F1 scores of 0.81 (internal) and 0.62 (external), with 72.7% of autonomous reports judged acceptable by board-certified radiologists, indicating readiness for limited deployment.
Supporting systems like MAViLT [6] and BiomedCLIP [3] demonstrate bidirectional report-image generation and improved retrieval, respectively, within constrained modalities.
Despite unprecedented progress, challenges remain. No current model effectively integrates all clinical modalities including EHR or waveform signals. Performance varies across modality pairs, and training such models requires extensive resources. Additionally, domain specialization often limits generalization, raising the need for more robust benchmarking across tasks and institutions.
Technical Challenges and Research Directions
Despite recent progress in integrating diffusion models with multimodal language systems for biomedical tasks, major obstacles remain across data, model design, and evaluation. This section outlines key challenges and proposes directions for future work.
Data Integration Challenges
Biomedical data is fragmented across institutions, modalities, and formats. While large datasets such as MIMIC-CXR [23] provide over 377,000 paired image–text examples, most diffusion-based approaches still train on narrow subsets (e.g., PathLDM [27] uses only TCGA-BRCA). Multimodal coverage is limited—datasets rarely contain triplets (e.g., CT, MRI, and report), unlike MedM2G [4] which unifies multiple imaging modalities.
Institution-specific biases, protocol drift, and equipment heterogeneity pose additional barriers. For instance, CXR-LLaVA [12] achieves an F1 score of 0.81 internally but drops to 0.62 on external validation, highlighting generalization gaps.
Privacy regulations such as HIPAA restrict centralized data sharing. Federated learning is a possible workaround [7], but models like Me-LLaMA [24], trained with 129 billion tokens and over 100,000 A100 GPU hours, remain infeasible in federated settings. Moreover, clinical reports often contain identifiable information, complicating de-identification and alignment for image-text pairs.
Finally, modality-specific representations remain a bottleneck. Histopathology images require high-resolution spatial tokenization, while radiology benefits from global context. Tokenization schemes such as VQ-GAN in MAViLT [6] and RadGraph-based embeddings in FactMM-RAG [20] remain hard to standardize across modalities.
Architectural Design Challenges
Unified models must balance scale with precision. MedM2G [4] handles high-resolution CT, MRI, and X-ray inputs simultaneously, incurring significant memory and training overhead. Me-LLaMA [24] demands over 100,000 A100 GPU hours—far beyond typical research budgets.
Clinical applications require pixel-level precision and explainability. While VQA-Diff [2] uses classifier guidance for question answering, its reasoning steps are not interpretable. Similarly, PathLDM [27] can generate realistic histopathology but lacks failure mode detection—making erroneous outputs difficult to catch.
Real-time constraints further complicate deployment. Diffusion-based models are inherently slower due to iterative sampling [8]. Emergency settings, such as trauma imaging or ICU monitoring, cannot afford long inference delays. Hospitals without high-end GPUs cannot deploy models like CXR-LLaVA [12] or ControlDiff [22] effectively. Streaming requirements also demand lightweight, continuous processing—an open problem for large multimodal systems.
Evaluation and Validation
Evaluation standards remain fragmented across tasks and modalities. Most benchmarks target isolated tasks: VQA-RAD for visual question answering, MIMIC-CXR for report generation, and SLAKE for short-answer VQA. Cross-modal benchmarks for joint synthesis or reasoning are rare, making holistic evaluation difficult [9]. Table 2 illustrates the diversity of current evaluation approaches and highlights key gaps in clinical validation.
| Challenge Category | Specific Issues | Current Limitations | Impact on Integration | Proposed Solutions |
|---|---|---|---|---|
| Benchmark Fragmentation | Task-specific metrics | VQA-RAD, MIMIC-CXR, SLAKE isolated | No cross-modal evaluation | Unified multimodal benchmarks |
| Performance Generalization | Institution bias | CXR-LLaVA: 0.81→0.62 F1 drop | Poor external validity | Cross-institutional validation |
| Clinical Acceptance | Below deployment threshold | 72.7% vs 84% required acceptance | Limited clinical adoption | Multi-reader validation protocols |
| Safety Validation | Hallucination detection | FactMM-RAG still produces errors | Patient safety risks | Uncertainty quantification [10] |
| Computational Feasibility | Resource requirements | Me-LLaMA: 100k+ A100 hours | Deployment barriers | Efficient architectures |
| Regulatory Compliance | FDA guidance gaps | No lifecycle management | Regulatory bottlenecks | Compliant validation frameworks |
| Interpretability | Black-box reasoning | VQA-Diff lacks explainability | Clinical trust issues | Intrinsic interpretability |
| Real-time Constraints | Inference latency | Diffusion sampling delays | Workflow integration failure | Optimized sampling strategies |
Table 2: Evaluation challenges and their impact on clinical integration of diffusion-MLLM systems
Clinical readiness requires more than accuracy. For instance, CXR-LLaVA [12] achieves 72.7% radiologist acceptance, but lags behind the 84% threshold typically required for autonomous clinical use. Multi-reader validation protocols and longitudinal assessments remain uncommon but are essential [21].
Safety and reliability are also underexplored. FactMM-RAG [20] improves factual correctness using retrieval augmentation, but still produces hallucinated findings. Domain bias still persists in large-scale models due to demographic skews or institutional labeling practices [3]. Regulatory requirements such as FDA guidance for AI-based medical devices emphasize lifecycle management, continuous validation, and explainability, yet few biomedical AI systems comply with them.
Future Work: We advocate for unified multimodal benchmarks, institution-agnostic validation pipelines, and interpretable training objectives. Integrating uncertainty quantification [10] and explainability into diffusion–LLM systems is a critical next step for responsible biomedical AI.
Future Directions for Integrated Diffusion–MLLM Systems
From Modular Coordination to Unified Architectures
Current Limitation: Our analysis reveals that existing systems like VQA-Diff [2] and FactMM-RAG [20] rely on shallow integration strategies—either treating vision and language independently or employing late fusion with minimal cross-modal interaction.
Research Directions:
- Shared latent spaces that unify visual and textual representations early in the model pipeline, extending beyond the domain-specific approaches in PathLDM [27]
- Joint optimization objectives that balance generation fidelity, reasoning accuracy, and clinical interpretability within a single training framework
- Bidirectional alignment schemes that enable seamless image-to-text and text-to-image generation, building on MAViLT’s [6] approach across diverse medical modalities
Scaling Across Modalities and Institutions
Current Limitation: Most systems demonstrate strong performance within narrow domains but face significant generalization challenges. CXR-LLaVA’s performance drop from F1 scores of 0.81 (internal) to 0.62 (external) [12] exemplifies this institutional bias problem.
Research Directions:
- Comprehensive foundation models trained across the full spectrum of biomedical data—imaging (CT, MRI, histopathology), text (clinical notes, literature), and structured data (genomics, lab values)
- Domain adaptation frameworks that maintain performance across diverse patient populations, institutional protocols, and equipment variations
- Privacy-preserving multimodal training through federated learning approaches [7, 10] that enable collaboration without centralizing sensitive medical data
Bridging Research Prototypes and Clinical Deployment
Current Limitation: Despite promising research results, significant barriers prevent clinical translation, including computational latency, lack of interpretability, and insufficient safety validation frameworks.
Research Directions:
- Real-time inference optimization through efficient sampling strategies and latent-space diffusion [18] to meet clinical workflow demands
- Intrinsic interpretability mechanisms that provide clinicians with transparent rationales, uncertainty estimates, and attention-based explanations integrated into the generation process
- Comprehensive evaluation frameworks that assess cross-modal consistency, clinical utility, and safety beyond traditional accuracy metrics [9]
- Continual learning capabilities that enable models to adapt to evolving medical knowledge and institutional protocols without catastrophic forgetting
Research Implementation Priorities
The transition from current modular approaches to truly integrated systems requires coordinated effort across multiple fronts. Immediate priorities should focus on developing shared tokenization standards and establishing multi-institutional evaluation consortiums. Medium-term goals include creating privacy-preserving training frameworks and demonstrating clinical utility in controlled settings. Long-term success will depend on sustained collaboration between AI researchers, clinicians, and regulatory bodies to ensure these integrated systems meet the stringent requirements of healthcare deployment.
Conclusion
This survey presents a comprehensive analysis of emerging strategies for integrating diffusion models with multimodal large language models (MLLMs) in biomedical AI. We introduced a taxonomy encompassing unified generative architectures, sequential pipelines, interleaved co-processing, and multi-agent frameworks, and systematically reviewed representative systems across core clinical tasks.
Our analysis highlights a structural gap between current modular implementations and the goal of unified, end-to-end biomedical reasoning systems. Despite substantial progress in both generative imaging and clinical language modeling, most systems exhibit shallow coupling, disjointed training regimes, and limited cross-modal alignment constraints that limit their scalability and clinical readiness.
To address these limitations, we identified key technical challenges related to data availability, architectural design, interpretability, and evaluation. We outlined research priorities including shared latent representations, co-optimized learning objectives, privacy-preserving training, and clinically grounded evaluation protocols.
The long-term goal is to develop integrated multimodal models capable of supporting transparent, adaptive, and clinically meaningful decision support across diverse biomedical contexts. Realizing this vision will require deeper architectural integration, rigorous validation, and sustained interdisciplinary collaboration. As the field advances, the frameworks and challenges outlined here may serve as a foundation for building next-generation biomedical AI systems that move beyond task-specific tools toward cohesive, clinically integrated intelligence.
References
[1] Alayrac, J. B., Donahue, J., Luc, P., Miech, A., Barr, I., Hasson, Y., … & Simonyan, K. (2022). Flamingo: a visual language model for few-shot learning. Advances in neural information processing systems, 35, 23716-23736.
[2] Bian, D., Wang, X., & Li, M. (2024, February). Diffusion-based Visual Representation Learning for Medical Question Answering. In Asian Conference on Machine Learning (pp. 169-184). PMLR.
[3] Boecking, B., Usuyama, N., Bannur, S., Castro, D. C., Schwaighofer, A., Hyland, S., … & Oktay, O. (2022, October). Making the most of text semantics to improve biomedical vision–language processing. In European conference on computer vision (pp. 1-21). Cham: Springer Nature Switzerland.
[4] Zhan, C., Lin, Y., Wang, G., Wang, H., & Wu, J. (2024). Medm2g: Unifying medical multi-modal generation via cross-guided diffusion with visual invariant. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 11502-11512).
[5] Dhariwal, P., & Nichol, A. (2021). Diffusion models beat gans on image synthesis. Advances in neural information processing systems, 34, 8780-8794.
[6] Evans, N., Baker, S., & Reed, M. (2025). A Generative Framework for Bidirectional Image-Report Understanding in Chest Radiography. arXiv preprint arXiv:2502.05926.
[7] Guan, H., Yap, P. T., Bozoki, A., & Liu, M. (2024). Federated learning for medical image analysis: A survey. Pattern Recognition, 110424.
[8] Ho, J., Jain, A., & Abbeel, P. (2020). Denoising diffusion probabilistic models. Advances in neural information processing systems, 33, 6840-6851.
[9] Kazerouni, A., Aghdam, E. K., Heidari, M., Azad, R., Fayyaz, M., Hacihaliloglu, I., & Merhof, D. (2023). Diffusion models in medical imaging: A comprehensive survey. Medical image analysis, 88, 102846.
[10] Koutsoubis, N., Waqas, A., Yilmaz, Y., Ramachandran, R. P., Schabath, M., & Rasool, G. (2024). Future-Proofing Medical Imaging with Privacy-Preserving Federated Learning and Uncertainty Quantification: A Review. arXiv preprint arXiv:2409.16340.
[11] Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240.
[12] Lee, S., Youn, J., Kim, H., Kim, M., & Yoon, S. H. (2025). CXR-LLAVA: a multimodal large language model for interpreting chest X-ray images. European Radiology, 1-13.
[13] Liu, H., Li, C., Wu, Q., & Lee, Y. J. (2023). Visual instruction tuning. Advances in neural information processing systems, 36, 34892-34916.
[14] Luo, R., Sun, L., Xia, Y., Qin, T., Zhang, S., Poon, H., & Liu, T. Y. (2022). BioGPT: generative pre-trained transformer for biomedical text generation and mining. Briefings in bioinformatics, 23(6), bbac409.
[15] Molino, D., di Feola, F., Shen, L., Soda, P., & Guarrasi, V. (2025). Any-to-Any Vision-Language Model for Multimodal X-ray Imaging and Radiological Report Generation. arXiv preprint arXiv:2505.01091.
[16] Nie, S., Zhu, F., You, Z., Zhang, X., Ou, J., Hu, J., … & Li, C. (2025). Large language diffusion models. arXiv preprint arXiv:2502.09992.
[17] Achiam, J., Adler, S., Agarwal, S., Ahmad, L., Akkaya, I., Aleman, F. L., … & McGrew, B. (2023). Gpt-4 technical report. arXiv preprint arXiv:2303.08774.
[18] Rombach, R., Blattmann, A., Lorenz, D., Esser, P., & Ommer, B. (2022). High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 10684-10695).
[19] Singhal, K., Azizi, S., Tu, T., Mahdavi, S. S., Wei, J., Chung, H. W., … & Natarajan, V. (2023). Large language models encode clinical knowledge. Nature, 620(7972), 172-180.
[20] Sun, L., Zhao, J., Han, M., & Xiong, C. (2024). Fact-aware multimodal retrieval augmentation for accurate medical radiology report generation. arXiv preprint arXiv:2407.15268.
[21] Tanno, R., Barrett, D. G., Sellergren, A., Ghaisas, S., Dathathri, S., See, A., … & Ktena, I. (2023). Consensus, dissensus and synergy between clinicians and specialist foundation models in radiology report generation. arXiv preprint arXiv:2311.18260.
[22] Tian, Y., Xia, F., & Song, Y. (2024, October). Diffusion Networks with Task-Specific Noise Control for Radiology Report Generation. In Proceedings of the 32nd ACM International Conference on Multimedia (pp. 1771-1780).
[23] Tiu, E., Talius, E., Patel, P., Langlotz, C. P., Ng, A. Y., & Rajpurkar, P. (2022). Expert-level detection of pathologies from unannotated chest X-ray images via self-supervised learning. Nature biomedical engineering, 6(12), 1399-1406.
[24] Xie, Q., Chen, Q., Chen, A., Peng, C., Hu, Y., Lin, F., … & Bian, J. (2024). Me-llama: Foundation large language models for medical applications. Research square, rs-3.
[25] Qin, J., Wu, J., Chen, W., Ren, Y., Li, H., Wu, H., Xiao, X., Wang, R., & Wen, S. (2024). DiffusionGPT: LLM-driven text-to-image generation system. arXiv preprint arXiv:2401.10061.
[26] Wu, S., Fei, H., Qu, L., Ji, W., & Chua, T. S. (2024, July). Next-gpt: Any-to-any multimodal llm. In Forty-first International Conference on Machine Learning.
[27] Yellapragada, S., Graikos, A., Prasanna, P., Kurc, T., Saltz, J., & Samaras, D. (2024). Pathldm: Text conditioned latent diffusion model for histopathology. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (pp. 5182-5191).
[28] Zhang, Z., Yao, L., Wang, B., Jha, D., Durak, G., Keles, E., … & Bagci, U. (2024). DiffBoost: Enhancing Medical Image Segmentation via Text-Guided Diffusion Model. IEEE Transactions on Medical Imaging.