Medical Image Segmentation
Medical image segmentation is an important component in the medical field, supporting the diagnosis of patients, treatment planning, and disease monitoring. Segmentation in machine learning is a process where datasets are broken into meaningful groups for annotation and deeper analysis. Medical segmentation, a combination of the two, has grown to importance in the field, but there still remains a challenging problem due to the large variability in modalities, anatomical structures, and usage. This report examines three recent approaches, MedSAM, UniverSeg, and GenSeg, that aim to address and improve on these limitations by improving generalization and adaptability.
Introduction
Medical segmentation plays a key role in decision making in clinical settings. It supports tasks ranging from diagnosis, treatment planning, and disease monitoring. However, the existence of large variability in imaging modalities, anatomical structures, and labeling conventions makes segmentation really difficult. In practice, medical images differ widely across different scanners, institutions, and patient populations.
Furthermore, new segmentation tasks come to light frequently that differ in both appearance and semantic definition from previous training data. These tasks may be centered on uncommon autonomies, institution specific labelling protocols, or generally unseen imaging conditions. This all makes it very difficult to design and scale medical image segmentation systems across diverse, real world clinical settings.
Motivation
Current existing approaches are catered towards narrowly defined tasks that still require a large amount of labelled data and retraining for new tasks. This leads to overeliance on very detailed task specific annotations or retraining continuously, both of which prove to be expensive and hard to maintain. As clinical environments continue to grow and evolve, these limitations hinder scalability and impact of medical image segmentation.
These challenges motivate the need for segmentation frameworks that are able to generalize well across new tasks in an efficient and effective manner. Ideally, these systems would support different formulations of task specification while simultaneously maintaining good performance across heterogeneous medical imaging domains. The following sections deep dive into three approaches that attempt to address these challenges from different yet complimentary perspectives.
MedSAM
Current medical segmentation models lack generalizability as many are tailored to individual medical image segmentation tasks. This can prove to be an obstacle for practical use of models in a clinical setting when there is a wide range of applications where medical segmentation can be applied. There already exist generalized segmentation models like the segment anything model (SAM) which is a general segmentation foundation model designed for general segmentation on natural images. However, when applied on medical image segmentations it performs conditionally with weaker performance when targets are characterized by weak boundaries causing a strong limitation of its use in the medical field. This led to the development of MedSAM [1], a promptable segmentation model specifically adapted for medical images. It can segment medical images both 2D and 3D(processes 3D images as a series of 2D slices) with the enhanced aid of specificity through user prompting.
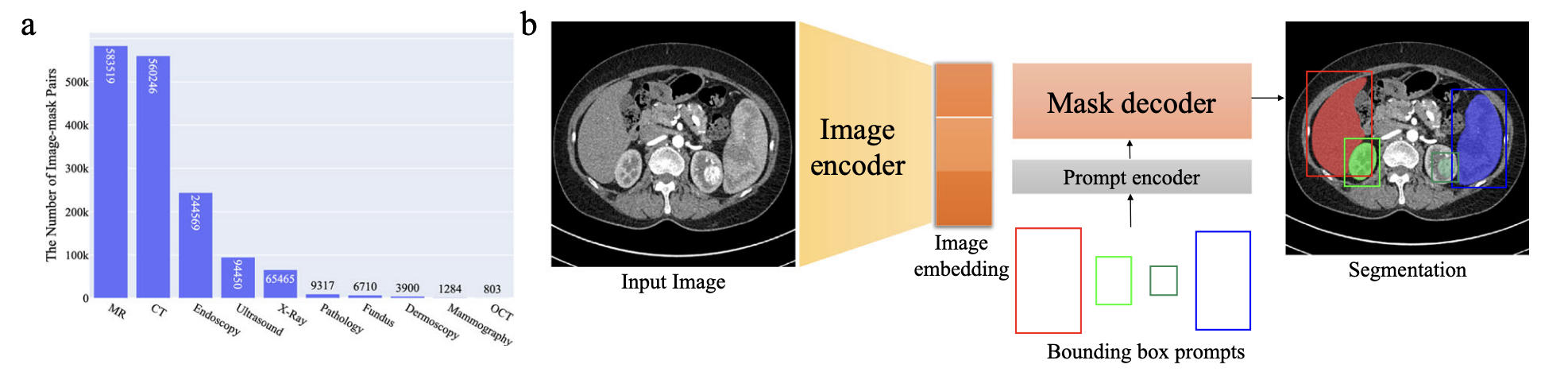
Fig 1. a. MedSAM dataset distribution b. MedSAM model architecture [1].
MedSAM’s approach was to finetune SAM on more than one million medical image mask pairs sourced from publicly available medical image segmentation datasets. To start, 1.5 million medical image mask pairs were split for training, tuning, and validation of the model. These medical images range from a variety of medical imaging modalities including CT, MRI, X-ray, ultrasound, endoscopy, ultrasound, mammography, OCT, and pathology and also cover a diverse range of targets like organs, tissues, and lesions to ensure generalization.
The data is then trained on a model retaining most of SAM’s backbone which includes a ViT-Base model with an image encoder that extracts dense visual features, a prompt encoder that processes user inputs, and a mask decoder that generates segmentation outputs. During training, bounding box prompts also help the model learn mapping by deriving ground truth masks which help convert coarse spatial localization to precise boundary delineation. This is aimed to help increase human interaction as in many medical settings, different clinicians may have different needs from the same image.
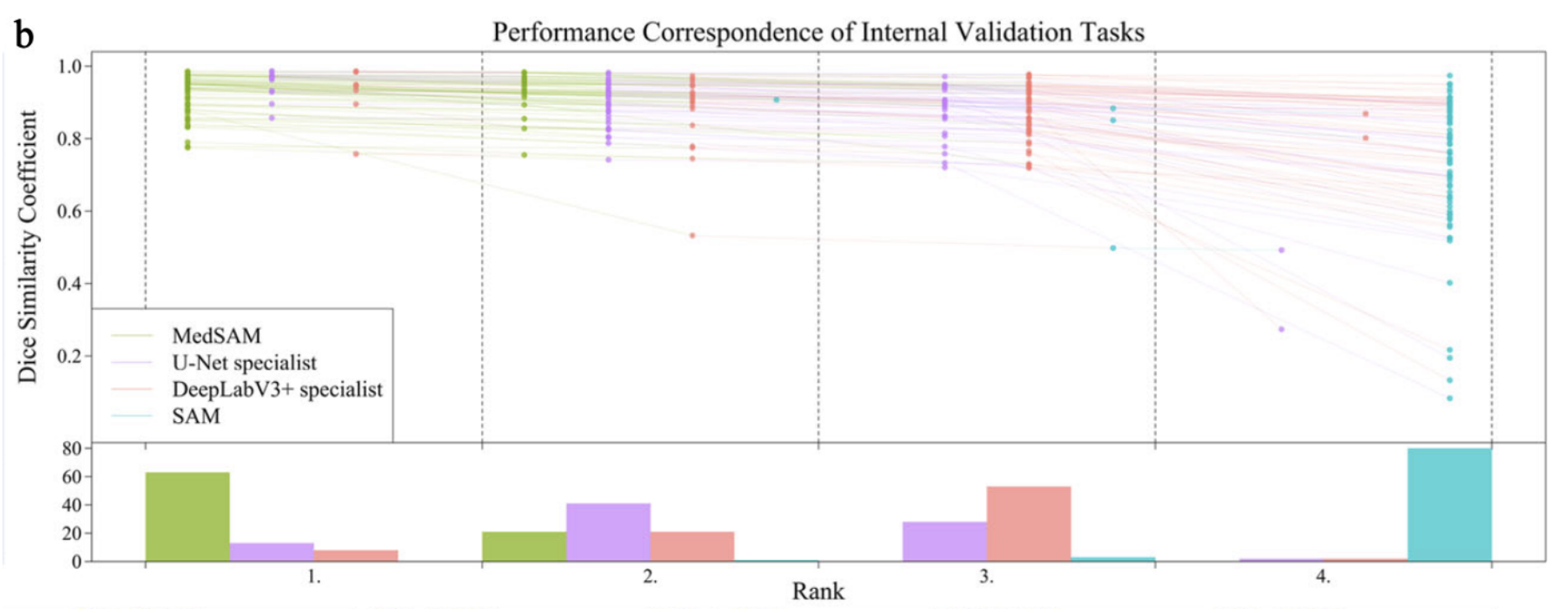
Fig 2. Performance Correspondence of Internal Validation Tasks across the models [1].
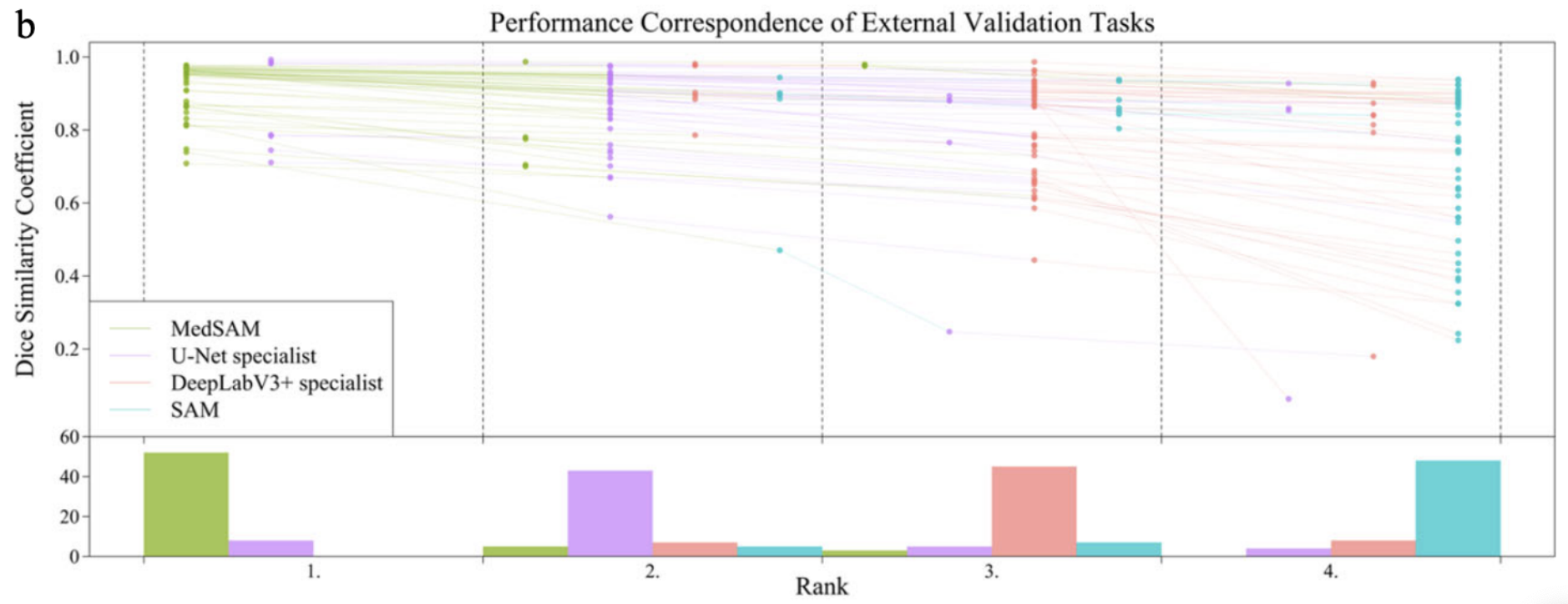
Fig 3. Performance Correspondence of External Validation Tasks across the models [1].
To evaluate the effectiveness of MedSAM, its performance was compared against SAM and specialist models like U-Net and DeepLabV3+. Each of the specialist models were trained separately for each of the modalities that MedSAM is trained on and then tested with internal and external validation where internal validation was the default data splits from the same dataset and the external validation came from unseen, new datasets of the same modality. Results then showed that MedSAM had a stronger consistent performance on internal and validation while U-Net and DeepLabV3+ would perform stronger on internal validation but poorly on external validation showing their limited generalization ability. SAM performed relatively poorly across all validation sets and in comparison to the other three models.
A human annotation study was also conducted to quantitatively evaluate MedSAM’s effect on annotation time cost in actual practice. Two experienced radiologists were given a dataset not in training or validation of the model to annotate through two pipelines: one with MedSAM and one without. To evaluate their annotations, the dice similarity coefficient and normalized surface distance (NSD) were.
\[DSC(G, S) = \frac{2|G \cap S|}{|G| + |S|}\]The dice similarity constant is a region based metric that measures the overlap between the two expert annotations and provides a quantitative statistic for segmentation accuracy.
\[NSD(G, S) = \frac{|\partial G \cap B_{\partial S}^{(\tau)}| + |\partial S \cap B_{\partial G}^{(\tau)}|}{|\partial G| + |\partial S|}\]NSD is a boundary based metric which evaluates how close the predicted annotations are within a specified tolerance parameter and this provides a measurement with more boundary precision. The combination of these two statistics capture the region level overlap and boundary level accuracy to comprehensively assess the use of MedSAM in actual practice, and they showed that the use of MedSAM can reduce annotation time by more than 80%.
In general, MedSAM is a strong step towards making segmentation foundation models useful in medicine and it shows that with large, well-curated medical data, a generalized segmentation model in medical uses is possible and broadly useful. However, there are still limitations to MedSAM. Much of the training data consist of CT, MRI, and endoscopy images which impact the models performance on other modalities like mammography. It is also limited on bounding box prompts for vessel-like branching structures. Overall, despite some limitations, MedSAM is still a significant improvement for medical image segmentation.
UniverSeg
The power of large scale foundational models for medical image segmentation is clearly demonstrated through MedSAM. However, the reality is that it still relies on predefined prompt formats to adapt to new segmentation issues and requires task specific fine tuning. UniverSeg [2] tackles the same ultimate goal of universality from a different and interesting angle. Instead of prompt engineering or retraining, it proposes an adaptation framework that is trained only once and is inference only. It essentially enables the segmentation of new medical tasks without additional learning.
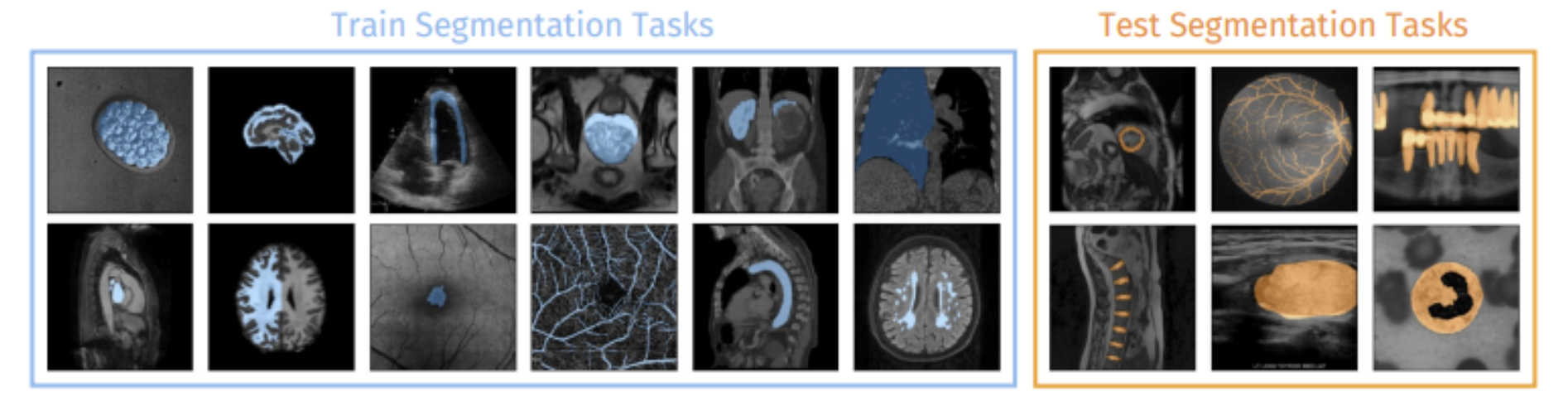
Fig 4. Train and Test Segmentation Tasks [2].
UniverSeg approaches medical image segmentation as a task conditioned inference problem. It does not encode the task implicitly using the weights of the model. Instead, the task is defined explicitly at inference time using a support set, which is a small collection of labelled image-mask pairs that represent the segmentation objective at hand. Given this support set and a query image, the model is able to output a segmentation for the query in a single forward pass. Essentially, if a few representative examples are given at inference time, UniverSeg is able to generalize to unseen anatomies, image modalities, and label spaces.
From an architectural perspective, UniverSeg contains a symmetric encoder that is responsible for processing both query and support images. This is then followed by a cross-feature interaction module, which is referred to as CrossConv or CrossBlock. This module is responsible for the facilitation of the bidirectional exchange of information between the query and support features. This essentially allows the model to understand what structures should be segmented in the query image based on the support examples available. Most importantly, this model does not assume that there are a fixed number of classes or predefined semantics. This makes it very well suited for heterogeneous and open set medical segmentation tasks.
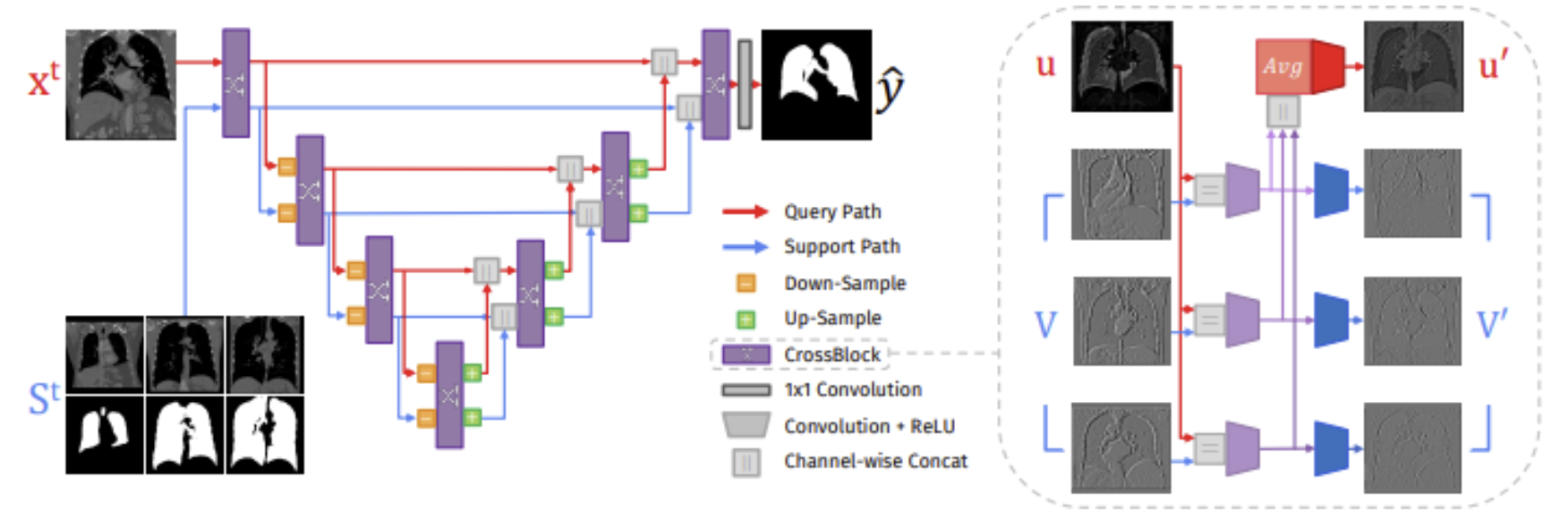
Fig 5. UniverSeg network (left) and CrossBlock architecture (right) [2].
The authors curated a very large and diverse training corpus named MegaMedical in order to train a model capable of broad generalization. MegaMedical consists of 53 public medical segmentation datasets. It spans various anatomical targets and imaging modalities (MRI, CT, fundus imaging, etc.). During the training process, segmentation tasks are sampled episodically. Each of the episodes define a new task through a randomly selected support set and the corresponding query images. This set up was designed to mirror the setting during inference. It really encourages the model to learn segmentation principles that are task agonistic instead of memorizing data specific patterns.
From an empirical standpoint, UniverSeg showcases both strong zero shot and few shot performance when tested on a large range of previously unseen data sets. The model was evaluated on segmentation tasks that differed greatly from the training distribution in terms of label definitions, autonomy, and modality. Furthermore, the results clearly show that UniverSeg outperforms traditional supervised baselines that have been trained on limited data. It is also competitive with or superior to several of the specialized few shot medical segmentation methods. It is crucial to note this performance is achieved without any fine tuning, truly highlighting the effectiveness of task conditioning through support examples.
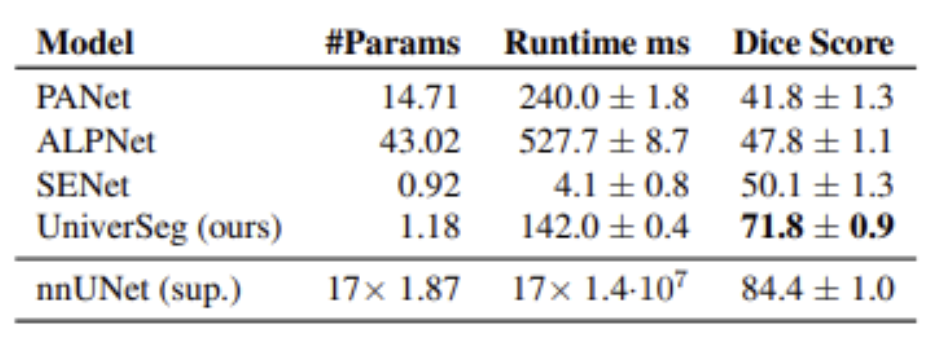
Table 1: Performance Summary of UniverSeg and each FS baseline (right) [2].
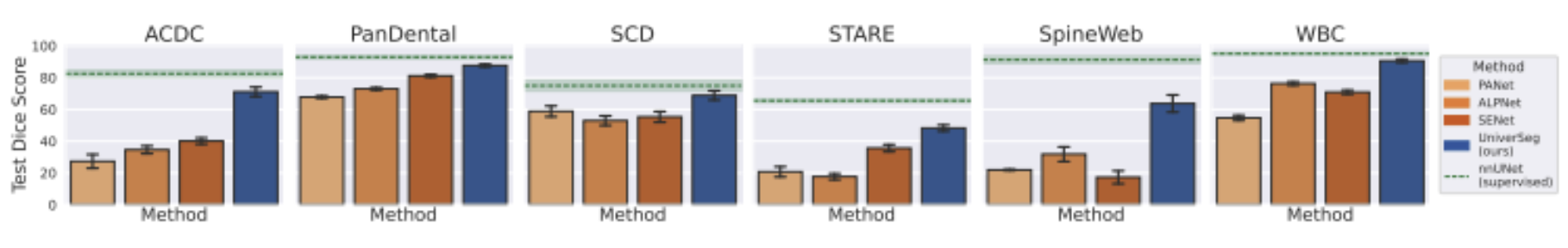
Fig 6. Average Dice score per each held out dataset [2].
While MedSAM focuses on promptable segmentation within a powerful model that has been pretrained, UniverSeg transfers the burden of adaptation from the parameters of the model to contextual examples. This represents an important design tradeoff in the realm of universal medical segmentation. MedSAM performs extremely well when simple prompts and large pretrained representations prove to be more than sufficient. On the other hand, UniverSeg truly shines in cases where the new tasks can not be encoded through prompts easily, labeled examples are present, and retraining is ineffective or inefficient. Together, these two approaches intertwine and provide interesting insights into complementary future work focusing on scalable and flexible medical image segmentation.
GenSeg
To complete our research of medical image segmentation, we read the paper, “Generative AI enables medical image segmentation in ultra low-data regimes.” In this paper, the authors aim to solve the problem of ultra low-data regimes. In the medical world, this refers to the extremely low availability of medical images that have been properly annotated due in part to limited professionals who can accurately complete the annotations, the time consuming nature of individually marking every pixel in an image, as well as the overall lack of adequate images for this annotation and analysis. To combat this issue, they introduce GenSeg: a framework used to generate synthetic data that has been labeled and can be used to train segmentation models [3].
Existing AI deep learning frameworks often struggle with medical segmentation due to their heavy reliance on large datasets. Approaches to augment and create synthetic data also often fail, as they treat augmentation as a separate step rather than integrating it with segmentation performance, which leads to inadequate data generation and training. Accurate segmentation performance is the critical aspect of medical imaging, yet the lack of incorporating this feedback is why recent approaches often struggle.
GenSeg, an end-to-end generative framework, is designed specifically to address the issue of the ultra low-data regimes. Its focus is to generate high-fidelity paired masks that can be used to train segmentation models more effectively. It differs from existing approaches by integrating data generation with segmentation performance. GenSeg generates these synthetic images by utilizing real medical image segmentation masks. The synthetic image-mask pairs are then used to train the model which in turn optimizes the generation. The framework uses a Generative Adversarial Network (GAN) based generator which consists of a generator and discriminator competing with each other, and builds upon this generator by embedding it inside its end-to-end framework. The feedback loop of this framework functions as follows: the generator produces the synthetic pair, the image-pair mask is then used to train a segmentation model, and the following performance is used as feedback to improve the generation process. The data generation is directly guided by the performance of the segmentation which ensures it is high-quality and accurate.
The authors were able to assess this improved accuracy by testing on as few as 40-100 training examples. As shown in Figure 7, the segmentation masks produced by GenSeg are a lot closer aligned with the Ground Truth than other popular models such as UNet. These results reveal that the generations produced are reliable and detailed even while being trained with very limited data.
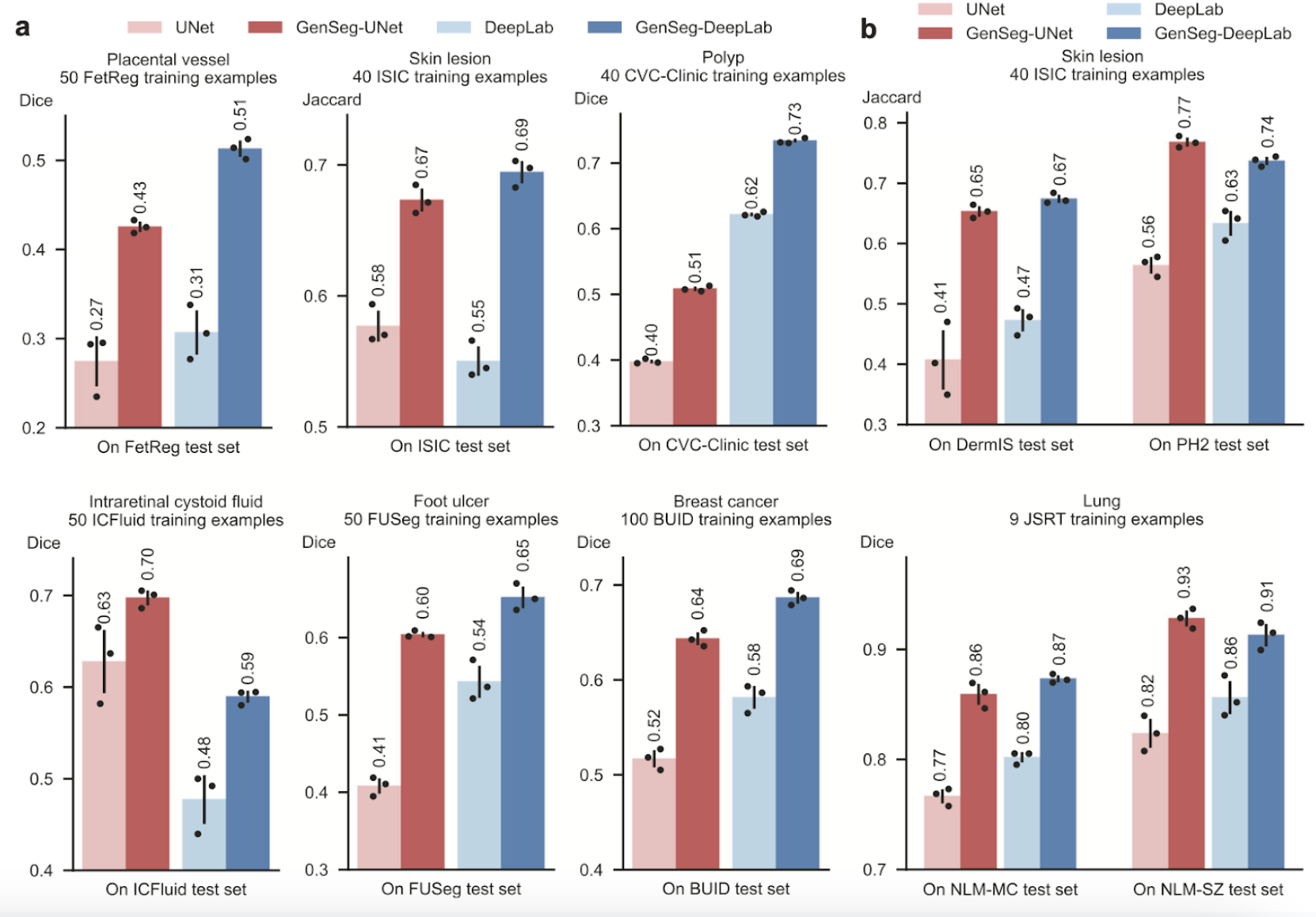
Fig 7. GenSeg performance comparison to Unet and DeepLab in ultra low-data regimes [3].
Additionally, the authors evaluated GenSeg in out-of-domain (OOD) scenarios. Out-of-domain refers to a situation where both the training and test data come from different datasets, and is common in the medical world when clinical data differs from training sets. GenSeg was able to outperform other models with under 40 labeled examples.
The authors were able to determine why GenSeg’s performance was so much more effective than existing approaches by examining components of the framework. The examinations showed that GenSeg yields higher segmentation in both in-domain and OOD settings which confirms that the multi-faceted approach of. Incorporating segmentation performance in data generation is the critical factor to improve the outcomes. The authors additionally conducted ablation studies to analyze individual components within the framework. These studies revealed that the end-to-end aspect of the framework is just as crucial as the joint segmentation and generation. Furthermore, the generator architecture improved performance with the adaptability in the generation approach based on segmentation performance. Another key finding is GenSeg does not need labeled data to outperform data augmentation techniques which is incredibly important to medical imaging with sensitive and private data.
The authors reveal that GenSeg is an effective solution that tackles the challenge of ultra low-data regimes of medical image segmentation. With the integration of generative modeling and segmentation performance in the end-to-end framework, GenSeg is able to reduce the need for large, annotated, trainable datasets while still achieving successful domain and OOD performance. The results show that data generation coupled with performance, rather than traditional augmentation, is the key to improving segmentation accuracy. Overall, GenSeg highlights the potential that generative AI has to meaningfully address limitations in the medical world.
Discussion
The three methods we have seen in the papers, MedSAM, UniverSeg, and GenSeg, all aim to improve generalization and scalability in medical image segmentation. MedSAM does this with its large-scale pretraining and is strong with enough data and prompts. UniverSeg builds upon this but conditions on support examples at inference without learning which is effective when tasks cannot be expressed with prompting. GenSeg encompasses all by addressing situations where there are neither prompts nor enough data present and focuses on improving performance with generation and segmentation performance.
The three approaches demonstrate constraints present in the medial imaging setting. MedSAM is best used for scenarios where prompts are available, likely interactive workflows. UniverSeg, in contrast, excels when adapting to new tasks given a small set of examples. GenSeg covers situations where there is less data present, particularly helpful for rare or un-understood diseases. Together, they demonstrate the different techniques to enhance medical imaging, and show that such approaches are complementary to combat a wide range of issues.
Conclusion
In conclusion, medical image segmentation remains to be a challenging yet necessary problem to solve. As these approaches reveal, ongoing research continues to improve the generalization and adaptability of segmentation frameworks in medical imaging. Each approach, MedSAM, UniverSeg, and GenSeg, tackles a key limitation, whether related to the availability of data, the use of prompts, or task adaptability, and together they highlight complementary strategies for improving medical image segmentation. Every new model and framework represents a step toward more accurate, scalable, and reliable analysis in an evolving clinical field.
Reference
[1] Ma, Jun, et al. “Segment anything in medical images.” Nature Communications, 2024.
[2] Butoi, Victor Ion, et al. “UniverSeg: Universal medical image segmentation.” International Conference on Computer Vision, 2023.
[3] Zhang, Li, et al. “Generative AI enables medical image segmentation in ultra low-data regimes.” Nature Communications, 2025.