An Introduction to Medical Image Segmentation
Medical image segmentation serves as the backbone of medical image processing in today’s world. In order to account for the variability in medical imaging, medical image segmentation detects boundaries within 2D and 3D images in order to identify crucial features and sizes of objects within them. This has tremendously assisted research, diagnosis, and computer-based surgery within the medical field. With the rise of deep learning algorithms, medical image segmentation has seen an increase in accuracy and performance and has led to incredible new innovations within the medical field.
- Introduction
- What is Medical Image Segmentation?
- U-Net
- Implementing a U-Net Model in Python
- Implementation for Brain Segmentation via U-Net
- Conclusion
- Video Demo
- Reference
Introduction
The rise of deep learning in recent years has allowed many fields of AI to flourish. Medical image segmentation existed before deep learning was around, but the introduction of deep learning led the resourcefulness and accuracy of such segmentation to flourish. There have been several main issues with medical image segmentation that previously limited the process’ abilities: variability within medical images, variability within human tissue, noise between image pixels, and inherent uncertainty due to limitations of knowledge within the medical world. Though these issues may remain indefinitely, deep learning has allowed the process of image segmentation to achieve better results than ever, and its potential is far more promising than the algorithms that came before it.
What is Medical Image Segmentation?
Medical image segmentation is the process of identifying main features within medical images by labeling each pixel with an object class such as ‘heart’, ‘tumor’, ‘artery’, etc. With deep learning, we are able to train models in order to automatically assign labels to pixels with high accuracy. These advancements have allowed the performance of automatic image segmentation to match that of professionally trained radiologists [2].
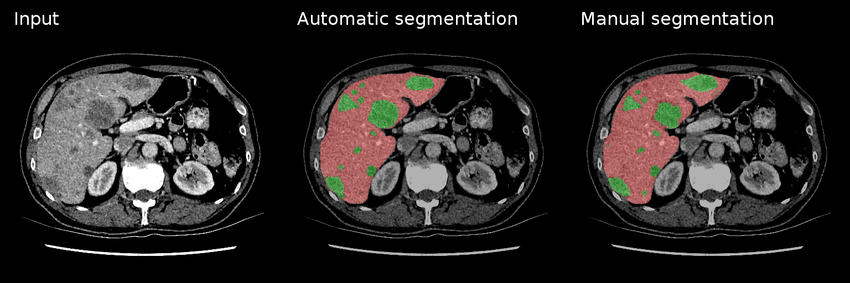
Fig 1. An example of manual & automatic segmentation on lesions (green) within a liver (red) [4].
As seen in Figure 1, the outputs of both manual and automatic segmentations of a liver with lesions are quite similar. And of course, the benefits of automatically segmenting these medical screenings include quicker interpretation times, less room for user error, and a second layer of analysis (first via the deep learning model, and second by a medical professional to confirm the accuracy of a scan’s results).
U-Net
U-Net is a type of convolutional neural network (CNN) used specifically for medical image segmentation. It is a very popular CNN due to its ability to train with fewer samples while still producing more accurate segmentations on testing data. It utilizes four encoders and four decoders which generates a U-shaped architecure, hence why the network got its name.
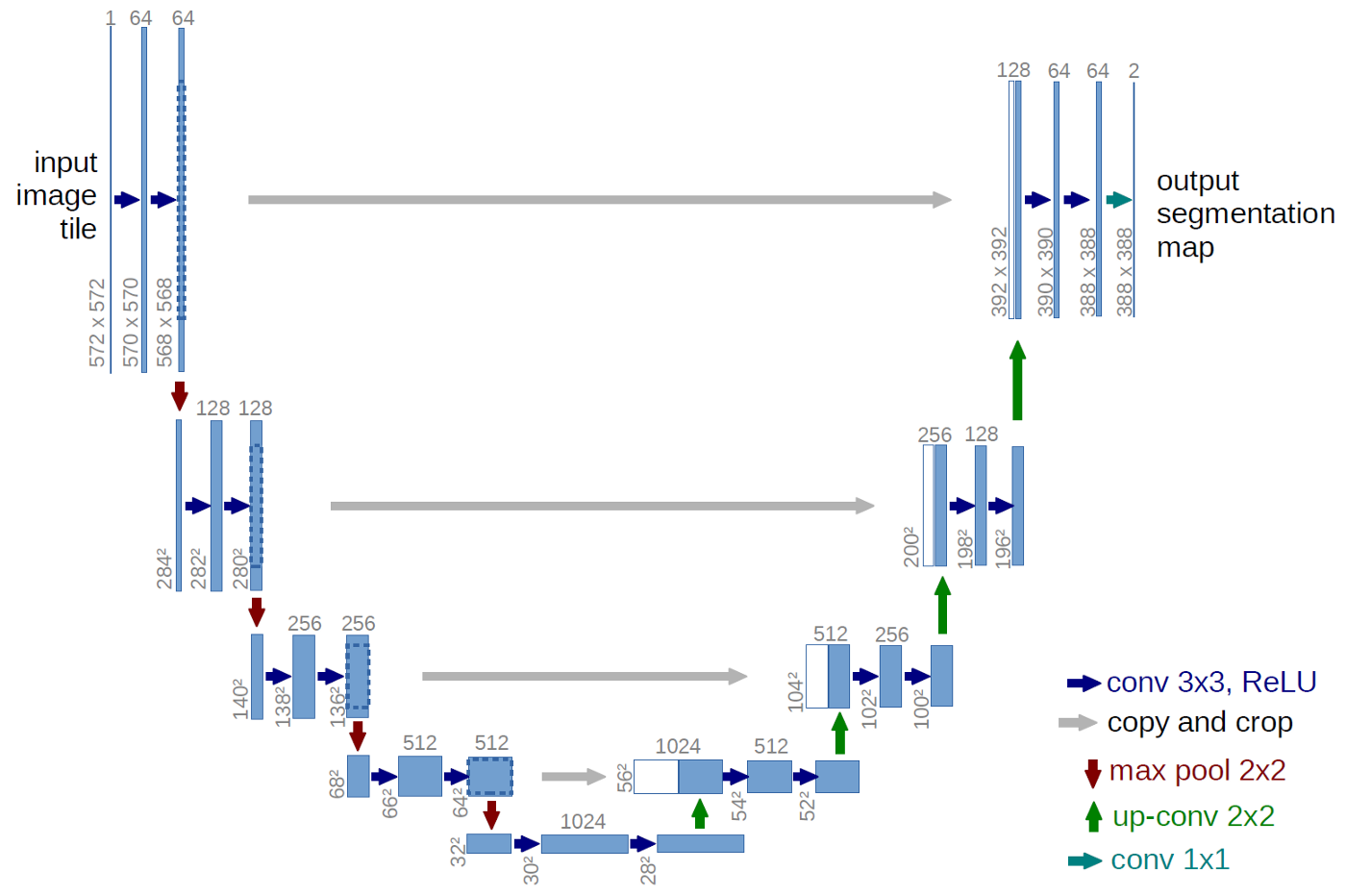
Fig 2. U-Net architecture consisting of four decoders, four encoders, and a bridge to connect the two. The blue boxes are feature maps, and the channel counts for each are listed above each box [5].
A simple implementation of what Figure 2 illustrates can be seen in Pytorch-UNet, a Python framework developed by the GitHub user milesial. It utilizes PyTorch to create encoder and decoder functions that process the features within each image and ultimately outputs a segmentation map which attempts to identify important characteristics within each image.
Encoding in U-Net
Encoders are networks that take an input and convert it to a feature map that identifies various important features within the original input. U-Net utilizes upsampling as its form of encoding which means each layer of encoding increases the output resolution. This allows high-resolution features to map more precisely following each convolution layer.
Within each encoder, we have two 3x3 convolutions followed by a Rectified Linear Unit (ReLU) activation function which helps prevent linearity as the model analyzes training data. After each block, we use a 2x2 max-pooling function which reduces the height & width of the feature map by 1/2 in order to maximize efficiency and minimize the cost of computation.
The encoder for U-Net resembles that of most convolutional neural networks [4]. Within each layer of downsampling described above, we double the number of features.
Decoding in U-Net
A decoder is used following the encoder and it includes upsampling–a process where artificially created data points are added to the dataset to restore the input image size–as well as concatenation of the upsampled path with the feature map from the encoder’s contracting path at that layer. The end result is the segmentation map used to identify features within the inputted image.
Achievements with U-Net
Various experiments have been conducted to test U-Net’s true capabilities in terms of accurate medical image segmentation. In a segmentation challenge on March 6, 2015, U-Net outperformed DIVE-SCI, IDSIA, and DIVE producing a warping error of 0.000353 (compared to a human error of 0.000005–the closest any CNN has gotten to mimicking human segmentation) [4]. It was also shown by the creators of MultiResUNet (a form of UNet that replaces sequencing two convolutional layers with a MultiRes block) that MultiResUNet performed with a very high accuracy rate showing that U-Net is very effective in processing images for segmentation.
Training a U-Net Model
As with any deep learning CNN model, we use training data and its corresponding output to train the model at hand. When compared with an error function of our choice, we determine how much a batch of training data should affect our model at each optimization step. A commonly used loss function is CrossEntropyLoss which has the following formula:
\[\mathbf{loss}(\it{x,class}) = -log\left(\frac{exp(\it{x}[\it{class}])}{\sum_{j} exp(x[j])}\right) = -\it{x}[\it{class}] + log\left(\sum_{j} exp(x[j])\right)\]The CrossEntropyLoss function calculates the difference of two probability distributions between a group of events. Its purpose is to change the weights of your model as you train it with your training data. When doing so, it adapts the model to more accurately classify or map the pixels to the final segmentation map. The lower the loss from the CrossEntropyLoss function, the better our model, so our goal is to minimize the loss as we train the model with our training data.
Implementing a U-Net Model in Python
Below is an example implementation of the U-Net model in Python. A version similar to this was initially created by PyTorch-Segmentation-Zoo on GitHub and was adapted for this article to resemble Figure 2 above. It shows how to create the model using PyTorch ‘nn’ functions in a simple class-based implementation. The U-Net class below utilizes the downSample class to execute the downsampling left-portion of the U-shape, while the upSample class executes the upsampling portion on the right side. Each class utilizes the double convolution function in order to execute the double 3x3 convolution.
def doubleConv(in_ch,out_ch):
sequential = nn.Sequential(
nn.Conv2d(in_ch,out_ch,3,padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True),
nn.Conv2d(out_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True)
)
return sequential
# Downsampling of U-Net
class downSample(nn.Module):
def __init__(self, in_ch, out_ch):
super(down_block,self).__init__()
self.maxpool = nn.MaxPool2d(2)
self.conv = doubleConv(in_ch,out_ch)
def forward(self, x):
x = self.maxpool(x)
x = self.conv(x)
return x
# Upsampling for U-Net
class upSample(nn.Module):
def __init__(self, in_ch, out_ch):
super(up_block,self).__init__()
self.up = nn.Upsample(scale_factor=2)
self.conv = doubleConv(in_ch,out_ch)
def forward(self, x1, x2):
x = self.up(x1)
x2_cropped = x2
x = torch.cat([x,x2_cropped],dim=1)
x = self.conv(x)
return x
class UNet(nn.Module):
def __init__(self, channels_in, classes_out):
super(UNet, self).__init__()
self.incoming = doubleConv(channels_in, 64)
self.down1 = downSample(64, 128)
self.down2 = downSample(128, 256)
self.down3 = downSample(256, 512)
self.down4 = downSample(512, 1024)
self.down5 = downSample(1024, 1024)
self.up1 = upSample(2048, 512)
self.up2 = upSample(1024, 256)
self.up3 = upSample(512, 128)
self.up4 = upSample(256, 64)
self.up5 = upSample(128, 64)
self.outcoming = nn.Conv2d(64,classes_out,1)
def forward(self, x):
x1 = self.incoming(x)
x2 = self.down1(x1)
x3 = self.down2(x2)
x4 = self.down3(x3)
x5 = self.down4(x4)
x6 = self.down5(x5)
x = self.up1(x6, x5)
x = self.up2(x, x4)
x = self.up3(x, x3)
x = self.up4(x, x2)
x = self.up5(x, x1)
x = self.outcoming(x)
return x
Implementation for Brain Segmentation via U-Net
A contributor to PyTorch by the GitHub username of mateuszbuda [5] created an open-source Python implementation of U-Net that allows you to implement medical image segmentation utilizing deep learning models and convolutional neural networks. Using this pretrained model, you can conduct medical image segmentation with just a few lines of code.
Setup
# Import necessary modules
import numpy as np
from PIL import Image
from torchvision import transforms
import torch
import urllib
# Import the pretrained model
model = torch.hub.load('mateuszbuda/brain-segmentation-pytorch', 'unet', in_channels=3, out_channels=1, init_features=32, pretrained=True)
# Gather an example image to feed to the model
url, filename = ("https://github.com/mateuszbuda/brain-segmentation-pytorch/raw/master/assets/TCGA_CS_4944.png", "TCGA_CS_4944.png")
try: urllib.URLopener().retrieve(url, filename)
except: urllib.request.urlretrieve(url, filename)
Conducting Image Segmentation
input_image = Image.open(filename)
m, s = np.mean(input_image, axis=(0, 1)), np.std(input_image, axis=(0, 1))
normalize = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize(mean=m, std=s),
])
input_tensor = normalize(input_image)
if torch.cuda.is_available():
input_batch = input_tensor.unsqueeze(0).cuda()
model = model.cuda()
with torch.no_grad():
output = model(input_batch)
print(torch.round(output[0]))
else:
print("Sorry... Must run with GPU!")
The output of the above block is a binary mask of size (1, H, W) that can be used to show the relevant segmentations of your image. Please reference the Brain Segmentation Google Colab Demo to see how you can visualize the resulting mask and utilize your segmentation map to conduct further medical examinations.
Conclusion
After examining the uses and one possible implementation of medical image segmentation, the benefits of such models should become much clearer. Medical image segmentation has allowed medical research to flourish, as automating the practice of segmenting images alleviates the many struggles of manual segmentation. We saw that U-Net, a type of convolutional neural network, excels in segmenting images, and open-source pretrained models make the setup process extremely easy to navigate. However, there are many other effective models out there such as DeepLab, SegNet, FirstGAN, SegAN, SCAN, and more. While this article only covers the possibilities of U-Net within medical image segmentation, there are many more studies that have been done covering a breadth of models within this field. Hopefully this article has opened your eyes to the exploding field of deep learning within medical research. With these new technologies in the medical field, we can expect many more innovations in the years to come.
Video Demo
Reference
[1] Müller, D., Kramer, F. MIScnn: a framework for medical image segmentation with convolutional neural networks and deep learning. BMC Med Imaging 21, 12 (2021). https://doi.org/10.1186/s12880-020-00543-7
[2] Liu, X.; Song, L.; Liu, S.; Zhang, Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021, 13, 1224. https://doi.org/10.3390/su13031224
[3] Vorontsov, Eugene & Tang, An & Pal, Chris & Kadoury, Samuel. (2018). Liver lesion segmentation informed by joint liver segmentation. 1332-1335. 10.1109/ISBI.2018.8363817.
[4] Olaf Ronneberger, Philipp Fischer, Thomas Brox. (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation
[5] Buda, Mateusz, Ashirbani Saha, en Maciej A. Mazurowski. “Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm”. Computers in Biology and Medicine 109 (2019): n. pag. Web.